The passing decade saw the viral spread of the Quick Response (QR) barcodes to every corner of our society. In China, near a billion of people now are using QR codes to pay restaurant bill, buy groceries, take a metro, or even make new friends via the phone. Such power comes from the digital transformation of our personal identity and other information to a tiny, black-white doted graph. Recently, a research team led by Prof. LI Jiong from Suzhou Institute of Nano-tech and Nano-bionics (SINANO) of Chinese Academy of Sciences (CAS) found a way to lever this power to detect many different biological signals at the molecular level and are aiming to apply the proposed tech platform for better clinical diagnostics.
Immunoassay is the primary method to quantitate molecules of interest from clinical specimens. Most immunoassays are only capable of measuring one specific molecular analyte in a run. The monoplex assay format is considered insufficient in analyte throughput, and low efficient as it causes multiplied cost of assay time, reagents, labor, and sample volume when constructing a panel test.
In their recently work published on Biosensors and Bioelectronics, Zheng et al. attempt to tackle this hurdle using a nanotechnology approach. They reported a multiplexed suspension array platform based on graphically encoded silica microparticles (Figure.1). The particles are in micron size and in planar shape.
Using lithography, researchers engraved even smaller, yet visible "dots" on the particle surface as the coding bits, resembling the form of a common QR code. A total of seven bits per particle provides a max coding space of 128-plex.
For parallel detection of multiple analytes, a selected group of coded particles are added to the sample. After a standard fluorescent immunoassay workflow, an automated microscope takes both the brightfield and fluorescent photos of the particles, which are analyzed by a computer program to decodes each particle and match the extracted signal intensity to it.
To evaluate this platform which they termed “GRASP assay”, the authors conducted parallel detection of up to six different cytokines in buffer solution. The analytical sensitivity reached subpicogram per ml level, while the dynamic range spans 4 logs (Figure.2).
These performances are superior to many commonly-used monoplex enzyme immunoassays (EIA) such as ELISA. The authors further demonstrated a clinical application by multiplex quantification of three auto-immune markers in serum from Type I diabetes patients. The sensitivity and specificity are equivalent to corresponding commercial diagnostic ELISA kits.
Compared to the widely-used bead cytometry multiplex assays depending on analog signals for decoding, the graphically-encoded GRASP assay provides a digital alternative which completely eliminates code mixing. It also avoids troublesome fluidics by using a standard multiwell plate as the assay vessel. Furthermore, the implement of semiconductor fabrication technology enables mass production of highly uniform particles in low cost.
For the next phase of their project, Dr. Li envisions to build a fully-automated in-vitro immunoassay system based on this technology in the near future.
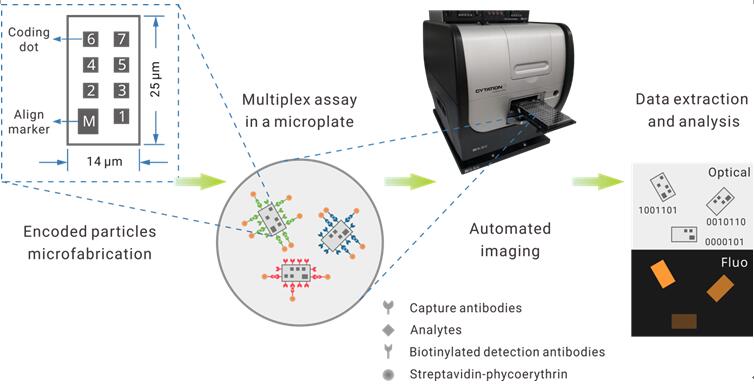
Figure.1 Design of the graphically-recognizable array of suspension particles (GRASP) assay.(Image by SINANO)
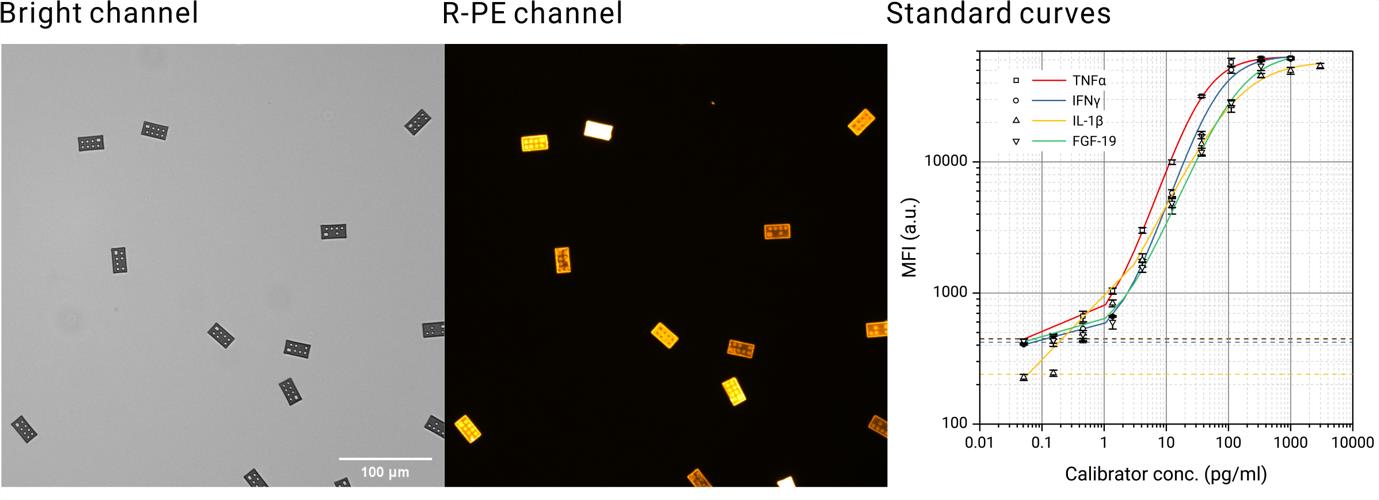
Figure.2 Particle images from a multiplex assay, and the obtained assay standard curves.(Image by SINANO)
Contact information: Prof.LI Jiong, Suzhou institute of Nano-Tech and Nano-Bionic, Chinese Academy of Sciences.
Email:jli2006@sinano.ac.cn
Reference:https://linkinghub.elsevier.com/retrieve/pii/S095656631930137X
downloadFile
