Small RNAs have emerged as one of the most important regulators for gene expression in diverse aspects of biology. Many kinds of small RNAs have been characterized, and these small RNAs are heterogeneous with respect to chemical modification. Some studies have indicated that microRNA (miRNA), small interfering RNA (siRNA), heterochromatic small interfering RNA (hc-siRNA), trans-acting siRNA (ta-siRNA) and natural antisense short interfering RNA (nat-siRNA) in plants, siRNA and Piwi-interacting RNA (piRNA) in insects, and piRNA in animals are 2'-O-methylated at the 3' end. Moreover, the 2'-O-methylation of small RNAs is functionally important for small RNA stability and function.
Although biologically crucial, 2'-O-methylation of small RNAs seems to be chemically trivial, especially for the purpose of profiling. Since the RNA sequence is by no means altered by the methylation, analytical biochemists could just assay them in the same way as they profiling the non-methylated species. Yet quite unexpectedly, in practice these tiny methyl groups have raised serious challenges for available high-throughput assay platforms (microarray and sequencing). As enzymatic labeling or ligation is an indispensable step for most commercial small RNAs profiling assays, these enzymes they used have to directly interact with the 3' end nucleotide of small RNAs where the methylation might hinder the catalytic reactions. Therefore, for 2'-O-methylation of small RNAs, the data produced by many enzyme-based assays are methylation-biased.
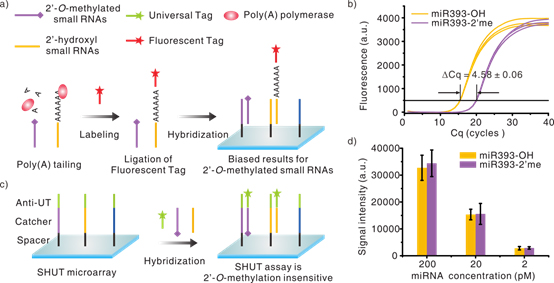
In 2011, Prof. LI Jiong’s group of Suzhou Institute of Nano-Tech and Nano-bionics (SINANO), CAS invented a novel label-free high-throughput small RNAs expression profiling (This work has been published in Nucleic Acids Research (click here to view paper)). Recently, they further demonstrate that this method is bias-proof to the end-nucleotide 2'-O-methylation of small RNAs. The method is label-free and enzyme-free, therefore born immune to the methylation-caused biases. The results show that the detection signal is correlated only to target concentration, no matter if the end nucleotide is modified or not (see Figure 1c and 1d). At the same time, they also evaluate the impact of 3' end 2'-O-methylation on the most commonly used enzyme Poly(A) polymerase. The results indicate that the methylated 3' end nucleotide is adenylated 23.9-fold less efficiently than the unmodified 3' end. Consequently, only about 4.2% of the methylated small RNAs may get labeled as compared to its non-methylated counterpart (see Figure 1a and 1b). Besides, this method inherits the advantages of high sensitivity (20 fM) and specificity (sharp discrimination of single-base mismatch near 5' end), and it requires only one hybridization step, allows direct use of total RNA without small RNA enrichment or labeling. In summary, this method first and successfully addressed the 2'-O-methylation-related detection biases for small RNAs, one of the most challenging and unsolved issues among current high-throughput small RNAs detection technologies. This work has been published in Analytical Chemistry (click here to view paper).
This work was supported by the Chinese Academy of Sciences, National Natural Science Foundation of China, and Natural Science Foundation of Jiangsu Province.
 |
|
Figure 1. (a) Poly(A) polymerase-based enzymatic labeling methods for small RNAs microarray (Affymetrix, Invitrogen, etc.).
(b) Quantification of 2′-O-methyl 3′end nucleotide bias using qPCR.
(c) Schematic representation of this method.
(d) The results of unmodified (miR393-OH) and 2′-O-methyl-modified small RNAs (miR393-2′me). |

