Hierarchical assemblies of biomolecules evolved in nature have been directed to inspire design and construction of artificial nanoscale devices. Particularly, engineering protein-DNA hybrids in a controlled manner is of great interest to nanotechnologists, arising from their important physiological roles and integrated molecular interactions which can bring new functionality and versatility for supramolecular assembly.
Notably, advances in DNA nanotechnology are impressive in recent years. Researchers have exploited a variety of complicate DNA structures from 1D to 3D via a range of rational design strategies based on the principle of complementary base pairing. These well-formed DNA scaffolds provide spatial addressability to arrange protein guests, and thus have enabled several applications including multi-enzyme cascades, nanoactuators, and as platforms for functional protein arrays. However, the methods for protein organization typically require chemical modification on protein surface, which is likely against the protein activity and render them inapplicable to in vivo systems. Examples of non-covalent conjugation of proteins on DNA scaffolds have also been demonstrated, by using biotin-streptavidin interaction, aptamers, or DNA binding proteins. Nevertheless, development of new strategies for in situ assembly of protein components with DNA is imperative especially with precise control over assembly orientation and protein-DNA stoichiometry.
Recently, a research team from Suzhou Institute of Nano-Tech and Nano-Bionics, Chinese Academy of Sciences successfully developed a facile strategy to construct controllable DNA origami–protein chimeric complexes, taking advantage of viral encapsidation mechanism and the programmability of DNA origami.
Viruses are typically composed of protein and nucleic acid, which can assemble rapidly in a highly organized fashion. The high-fidelity of virus packaging procedure reveals specific interactions between coat proteins and its genomic nucleic acid. Based on previous research on virus-like particle assembly and DNA nanotechnology, the Qiangbin Wang's group employs locally driven in situ assembly of tobacco mosaic virus (TMV) coat proteins onto DNA origami. The TMV genome-mimicking RNA strands containing a characteristic loop of the origin of assembly sequence were selectively immobilized on the surface of DNA origami scaffold. The engineered RNA strands with different lengths were designed to tailor protein nanotube lengths and enable refined stoichiometry control of protein components in a defined growth direction. Thus, the polarized in situ assembly process on DNA scaffold can be achieved and resulted in versatile DNA origami-protein hierarchical nanoarchitectures with precisely prescribed configurations.
This strategy can be applied to other virus systems for the construction of multi-component complexes. DNA nanostructure provides higher spatial precision for custom arrangement of viral proteins. The controllability of virus-based functional materials can be improved upon the usage of viral-like particles positioned in these hybrid assemblies, thereby generating novel properties. Moreover, the highly repetitive coat proteins of virus are good carrier to fuse multiple peptides or proteins by genetic engineering, which may largely benefit hybrid system both in structural complexity and functionality.
This work entitled "Selective in Situ Assembly of Viral Protein onto DNA Origami" was recently published in Journal of the American Chemical Society. The work was supported by grants from the National Natural Science Foundation of China and the National Key Research and Development Program.
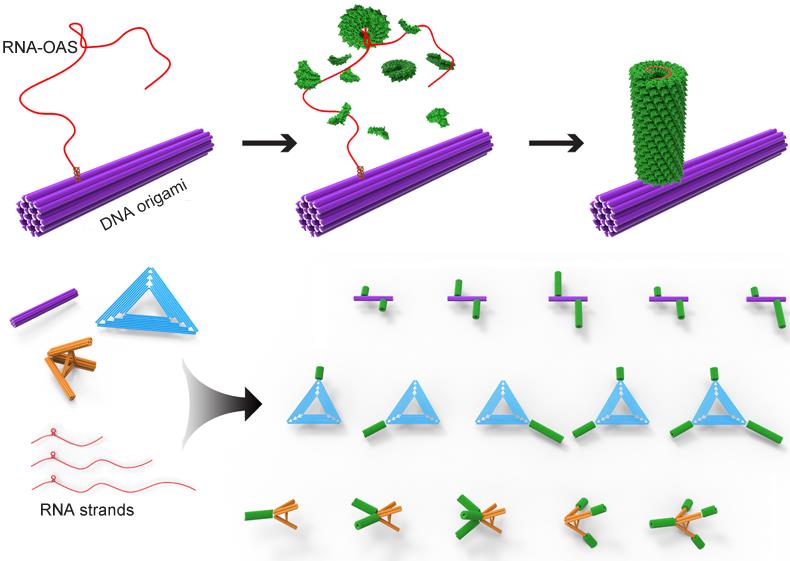
Figure1 Schematic illustration of the in situ assembly of a protein nanotube onto a rigid 24HB DNA origami and the formation of versatile DNA origami-protein hierarchical nanoarchitectures(Image by SINANO).
Contact information:
Prof.WANG Qiangbin, Suzhou institute of Nano-Tech and Nano-Bionic, Chinese Academy of Sciences.
Email:qbwang2008@sinano.ac.cn
Reference:https://pubs.acs.org/doi/10.1021/jacs.8b03914

